Prevalence, Genome Characterization, and Phylogenetic Analysis of Bovine Hepacivirus in Inner Mongolia, Northeastern China
# contributed equally to this work, * Corresponding author
Abstract
Objective: Bovine hepacivirus (BovHepV) is a new member of the genus Hepacivirus in the family Flaviviridae, which has been detected in cattle in more than seven countries. The purpose of the study was to identify and genetically characterize BovHepV in cattle in northeastern China.
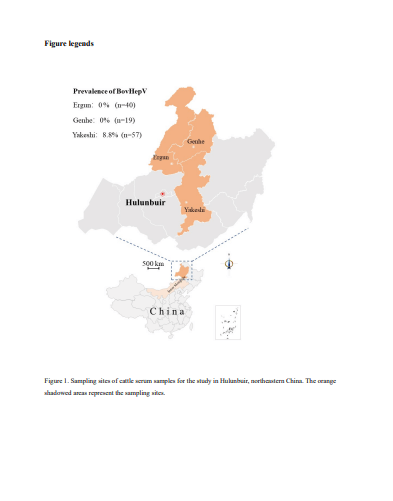
Methods: A total of 116 serum samples from free-range cattle were collected from HulunBuir city in April and May, 2021, and were divided into three pools before being subjected to metagenomic sequencing. The BovHepV sequences obtained were used for genome characterization and phylogenetic analysis.
Results: Two 8840 nucleotides long BovHepV strains YKS01/02 were identified in the samples through metagenomic sequencing. In RT-PCR screening, the prevalence of BovHepV was 8.8% (5/57) in Yakeshi of HulunBuir. The novel virus showed a range of 79.3%–91.9% nucleotide sequence identity and 92.5%–98.1% amino acid sequence identity with the BovHepV sequences discovered. Phylogenetic analysis classified YKS01/02 strains into BovHepV subtype G strains discovered in Jiangsu Province, China, and named with subtype G2.
Conclusion: This study first identified BovHepV in cattle in northeastern China, and expands the geographical distribution and genetic diversity of BovHepV in the country.
- 04 Apr 2022 15:39 Version 1
Rapid Rating Times:
4
· Level of
Quality: 4.25· Level of Repeatability: 4.25
· Level of Innovation: 3.25
· Level of Impact: 3.5
*Each rating ranges from 0-5
-
Level of QualityIs the publication of relevance for the academic community and does it provide important insights? Is the language correct and easy to understand for an academic in the field? Are the figures well displayed and captions properly described? Is the article systematically and logically organized?0.0
-
Level of RepeatabilityIs the hypothesis clearly formulated? Is the argumentation stringent? Are the data sound, well-controlled and statistically significant? Is the interpretation balanced and supported by the data? Are appropriate and state-of-the-art methods used?0.0
-
Level of InnovationDoes the work represent a novel approach or new findings in comparison with other publications in the field?0.0
-
Level of ImpactDoes the work have potential huge impact to the related research area?0.0
-
Abstracts1447
-
PDF Downloads910